Description
Product Description
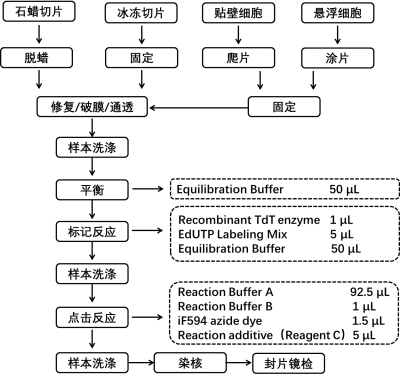
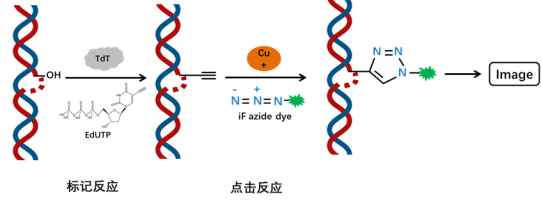
The fragmentation of chromosomal DNA during cellular apoptosis is a progressive and sequential process. Initially, chromosomal DNA is degraded into large fragments of 50-300 kb by endogenous nucleases. Subsequently, approximately 30% of the chromosomal DNA is randomly cleaved between nucleosome units by Ca2+ and Mg2+-dependent endonucleases, resulting in the formation of 180-200 bp nucleosomal DNA polymers. Therefore, during the late stages of cellular apoptosis, DNA is degraded into fragments of 180-200 bp, exposing a significant amount of 3′-OH ends on the fragmented genomic DNA. Terminal deoxynucleotidyl transferase (TdT) is a template-independent DNA polymerase that catalyzes the addition of deoxynucleotides to the 3′-OH ends of fragmented DNA molecules. Hence, the TUNEL (TdT-mediated dUTP nick end labeling) assay kit can be used to detect the fragmentation of nuclear DNA in tissue cells during the late stages of apoptosis.
The principle of the assay is based on the incorporation of EdUTP (a modified dUTP with an alkyne group) into the exposed 3′-OH ends of genomic DNA during DNA fragmentation, catalyzed by TdT enzyme. The alkyne group then undergoes a click reaction with an azide dye (iF594 azide) in the presence of monovalent copper ions, resulting in the site-specific introduction of a fluorescent group. The detection can be performed using a fluorescence microscope or flow cytometer (iF594 excitation at 593 nm and emission at 614 nm). Compared to other modified dUTPs, EdUTP has smaller steric hindrance, making it more accessible for incorporation by TdT enzyme at DNA ends.
This assay kit has a wide range of applications and is suitable for the detection of cellular apoptosis in various samples, including paraffin-embedded tissue sections, frozen tissue sections, cell smears, and cell monolayers.
Storage and Transportation
The product should be transported with wet ice (ice packs) and stored at -20°C. It has a shelf life of 12 months.
| Component Number | Component | G1516-50T |
| G1516-1 | Recombinant TdT Enzyme | 50 µL |
| G1516-2 | EdUTP Labeling Mix | 250 µL |
| G1516-3 | Equilibration Buffer | 5×1 mL |
| G1516-4 | Proteinase K(200 µg/mL) | 1 mL |
| G1516-5 | iF594 azide dye | 80 μL |
| G1516-6 | Reaction Buffer A | 5×1 mL |
| G1516-7 | Reaction Buffer B | 60 μL |
| G1516-8 | Reaction additive(Reagent C) | 2×100 mg |
| Instruction | 1 | |
Preparation Before Experiment
- PBS phosphate buffer solution (recommended G0002 or G4202).
- Fixation solution: 4% paraformaldehyde dissolved in PBS, pH 7.4 (recommended G1101).
- Permeabilization solution: 0.1% Triton X-100 dissolved in 0.1% sodium citrate (recommended G1204).
- 0.2% Triton X-100 in PBS; 0.1% Triton X-100 in PBS containing 5 mg/mL BSA.
- If nuclear staining is required, prepare DAPI (2 µg/mL), Hoechst 33258, or PI (1 µg/mL) separately (recommended G1012, G1011, G1021).
- If a positive control experiment is needed, prepare DNase I separately (recommended G3342).
- If using a flow cytometer, prepare PI staining solution (recommended G1021) and RNase A (DNase-free) (recommended G3413).
- Reaction additive (Reagent C): Dissolve 100 mg powder in 1 mL ultrapure water (ready for use), aliquot 100 μL, and store at -20°C. Keep the remaining powder for future use. (Reagent C is prone to oxidation, so avoid prolonged exposure to air. It is strongly recommended to aliquot the solution into smaller portions after dissolution. It has been tested that slight color changes in the Reagent C solution do not affect the performance of the click reaction system. If the solution turns brown, it indicates the component has become ineffective.)
- If the background color of the results is too dark, it may be due to insufficient washing or residual fixation solution during the experiment.
- Pay attention to the sequential addition of reagents in the click reaction system, and mix well while adding.
- For your safety and health, please wear a lab coat and disposable gloves while conducting the experiment.
Step 1: Sample Preparation
A. Paraffin-embedded tissue sections
- Immerse the paraffin tissue sections in xylene at room temperature for 5-10 minutes, repeating 2-3 times.
- Immerse the sections in absolute ethanol for 5 minutes, repeating 2 times.
- Immerse the sections in gradient ethanol (85%, 75%, and double-distilled water) once each for 5 minutes.
- Gently rinse the sections with PBS and remove excess liquid. Use a histology pen to draw a small circle about 2-3 mm away from the tissue, allowing for subsequent permeabilization and labeling steps. Avoid letting the samples dry out during the experiment. Keep the processed samples in a humid chamber.
- Prepare Proteinase K working solution by diluting Proteinase K (200 µg/mL) stock solution with PBS in a 1:9 ratio to achieve a final concentration of 20 μg/mL.
- Add 100 μL of the Proteinase K working solution to each sample, ensuring complete coverage, and incubate at 37°C for 20 minutes.
- Wash the samples 3 times with PBS for 5 minutes each (thoroughly rinse off Proteinase K, as it may interfere with subsequent labeling reactions). Keep the processed samples in a humid chamber.
- (Optional step) Remove excess liquid from the samples and add an appropriate amount of membrane permeabilization solution to fully infiltrate the tissue. Incubate at room temperature for 20 minutes. After membrane permeabilization, wash the samples 3 times with PBS for 5 minutes each. Keep the processed samples in a humid chamber.
B. Frozen tissue sections
- Immerse the tissue sections in a fixative solution and incubate at room temperature for 10-15 minutes.
- Remove the tissue sections from the fixative solution and let them air dry in a fume hood.
- Rinse the tissue sections in distilled water or PBS to remove residual fixative.
- Use a histology pen to draw a small circle about 2-3 mm away from the tissue, allowing for subsequent permeabilization and labeling steps. Avoid letting the samples dry out during the experiment. Keep the processed samples in a humid chamber.
- Prepare Proteinase K working solution by diluting Proteinase K (200 µg/mL) stock solution with PBS in a 1:9 ratio to achieve a final concentration of 20 μg/mL.
- Add 100 μL of the Proteinase K working solution to each sample, ensuring complete coverage, and incubate at room temperature for 10 minutes.
- Wash the samples 2-3 times with PBS to remove excess liquid (thoroughly rinse off Proteinase K, as it may interfere with subsequent labeling reactions). Keep the processed samples in a humid chamber.
- (Optional step) Add an appropriate amount of membrane permeabilization solution to fully infiltrate the tissue. Incubate at room temperature for 20 minutes. After membrane permeabilization, wash the samples with PBS to remove excess liquid. Keep the processed samples in a humid chamber.
C. Cell climbing slides
- Culture adherent cells on Lab-Tek chamber slides. After induction of apoptosis, gently rinse the chamber slides twice with PBS.
- Add an appropriate amount of fixative solution to cover the tissue in each chamber slide and incubate at room temperature for 20 minutes.
- Remove the fixative solution and wash the slides with PBS three times for 5 minutes each.
- Immerse each sample in a 0.2% Triton X-100 solution for 5 minutes to permeabilize the cell membranes.
- Wash the slides three times with PBS for 5 minutes each.
- Proceed to the labeling step according to your experiment’s requirements.
Please note that this translation may not be perfect, but it should give you a general understanding of the steps.
| Component | Volume |
| Reaction Buffer A | 925 μL |
| Reaction Buffer B | 10 μL |
| iF594 azide dye | 15 μL |
| Reaction additive(Reagent C) | 50 μL |
| total | 1000 μL |
- Remove the reaction solution by clicking and immediately wash it 2-3 times with PBS buffer, 5 minutes each time.
- Gently remove the PBS solution around the sample using filter paper.
- Nuclear staining: Stain the sample in a staining dish by immersing the glass slide in the staining dish containing DAPI solution (prepared and diluted with fresh PBS) in the dark, and leave it at room temperature for 8 minutes (or use Hoechst 33258 for nuclear staining).
- Coverslipping: After the sample is stained, wash the tissue sample three times with PBS, 5 minutes each time. Then gently remove the excess liquid and add anti-fade mounting medium (recommended: G1401) for coverslipping.
- Microscopic examination: Analyze the sample immediately under a fluorescence microscope. Handle the glass slide with caution and avoid light exposure. DAPI stains both apoptotic and non-apoptotic cells blue, while the iF594 azide dye is only incorporated into the apoptotic cell nuclei, resulting in red fluorescence.
IV. Detection of Suspended Cells Using Flow Cytometry
- Wash the cells to be tested twice with PBS, centrifuge at 4°C (500 g), and then resuspend in 500 µL of PBS.
- Fixation: Add 5 mL of 1% paraformaldehyde solution prepared with PBS to the sample to fix the cells. Place on ice for 20 minutes.
- Centrifuge the cells at 4°C, 300 g for 10 minutes, remove the supernatant, and resuspend with 5 mL of PBS twice. Finally, resuspend the cells in 500 µL of PBS.
- Permeabilization: Add 0.2% Triton X-100 solution prepared with PBS to the cell sample for permeabilization. Leave at room temperature for 5-10 minutes.
- Centrifuge the cells at 300 g for 10 minutes, resuspend with 5 mL of PBS, and centrifuge again. Then, resuspend with 1 mL of PBS.
- Equilibration: Transfer about 2×10^6 cells to a 1.5 mL microcentrifuge tube, centrifuge at 300 g for 10 minutes, remove the supernatant, and resuspend with 80 μL of Equilibration Buffer. Incubate at room temperature for 5 minutes.
- Labeling solution preparation: Thaw EdUTP Labeling Mix and Equilibration Buffer on ice, and mix Recombinant TdT enzyme, EdUTP Labeling Mix, and Equilibration Buffer in a ratio of 1 μL:5 μL:50 μL (1:5:50) to prepare enough TdT incubation buffer for all experiments and optional positive control reactions.
- Labeling: Centrifuge the cells at 300 g for 10 minutes, remove the supernatant, and resuspend the pellet in 56 μL of TdT incubation buffer. Incubate at 37°C for 1 hour, avoiding light. Gently resuspend the cells every 15 minutes using a micropipette.
- After the reaction is complete, remove the labeling reaction solution and wash the sample with PBS, repeating the wash step 3-4 times, 5 minutes each time.
- Click reaction: Remove the previous PBS buffer and add 100 μL of click reaction solution onto the sample, ensuring it covers the entire sample. Incubate at room temperature, avoiding light, for 30 minutes. (Refer to the table below for the composition of the click reaction solution. Add each reagent sequentially while mixing. The proportions can be adjusted proportionally. It is recommended to prepare in advance.)
Please note that the table mentioned in step 10 is missing, so I couldn’t provide the specific composition of the click reaction solution.
| Component | Volume |
| Reaction Buffer A | 925 μL |
| Reaction Buffer B | 10 μL |
| iF594 azide dye | 15 μL |
| Reaction additive(Reagent C) | 50 μL |
| 总体积 | 1000 μL |
- Remove the click reaction solution and immediately wash it 2-3 times with PBS buffer, 5 minutes each time.
- Remove the supernatant and resuspend the pellet in 1 mL of 0.1% Triton X-100 solution prepared with PBS, which contains 5 mg/mL BSA. Repeat the wash step 2 times.
- Nuclear staining: Centrifuge at 300 g for 10 minutes, remove the supernatant, and resuspend the cell pellet in 0.5 mL of DAPI solution containing 250 μg of RNase A without DNase. Incubate the cells in the dark at room temperature for 30 minutes (or use Hoechst 33258 for nuclear staining).
- Analysis: Analyze the cells using a flow cytometer. DAPI stains both apoptotic and non-apoptotic cells blue, while the iF594 azide dye is only incorporated into the apoptotic cell nuclei, resulting in red fluorescence.