Description
Product Information
| Product Name | Cat.No. | Spec. |
| 2 × SYBR Green qPCR Master Mix (High ROX) | G3322-01 | 1 mL |
| G3322-05 | 5×1 mL | |
| G3322-15 | 15×1 mL |
Description/ Introduction
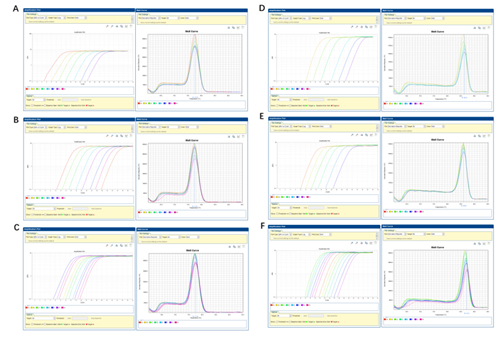
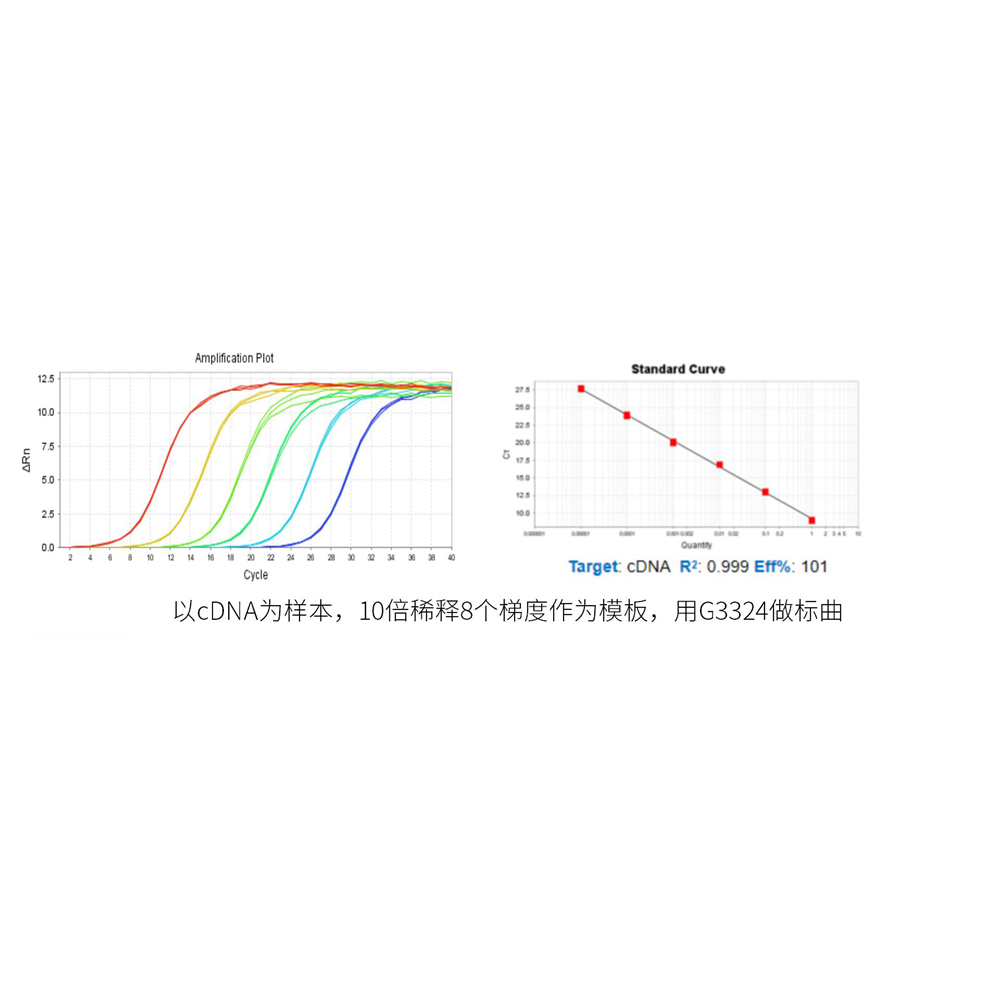
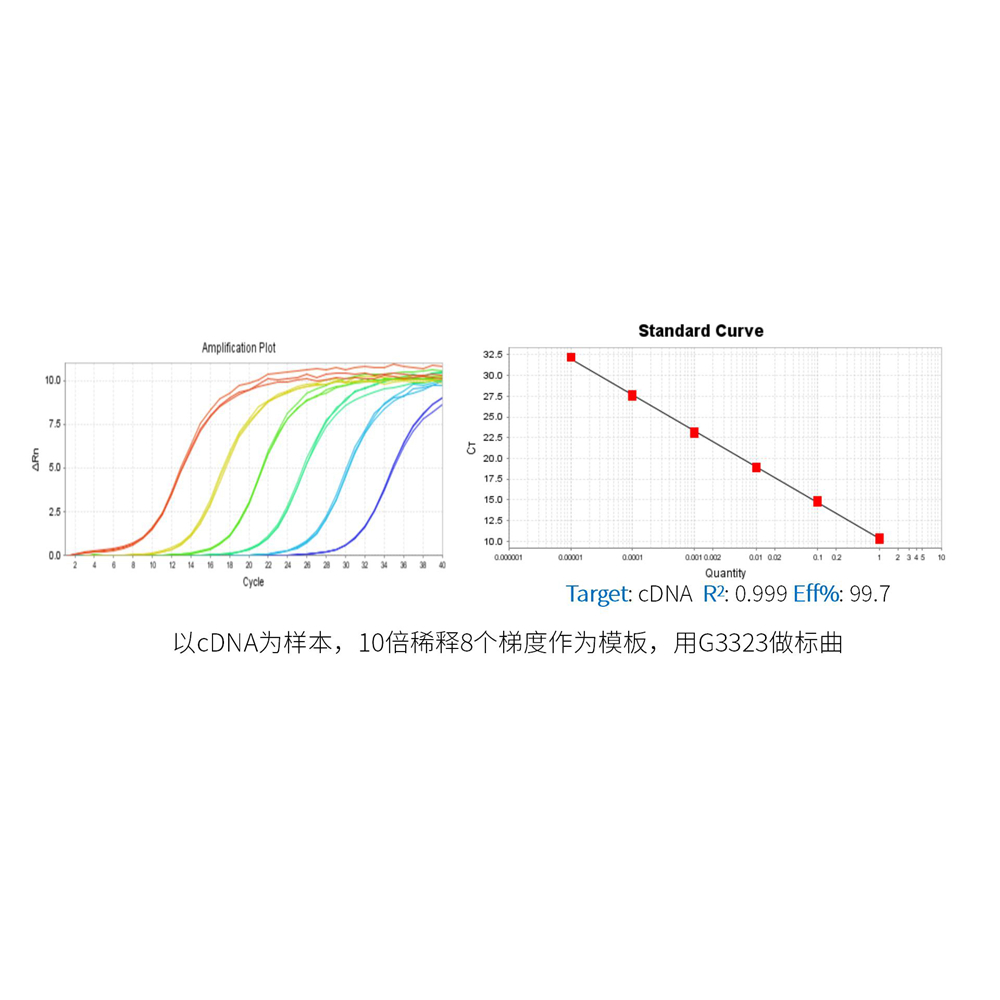
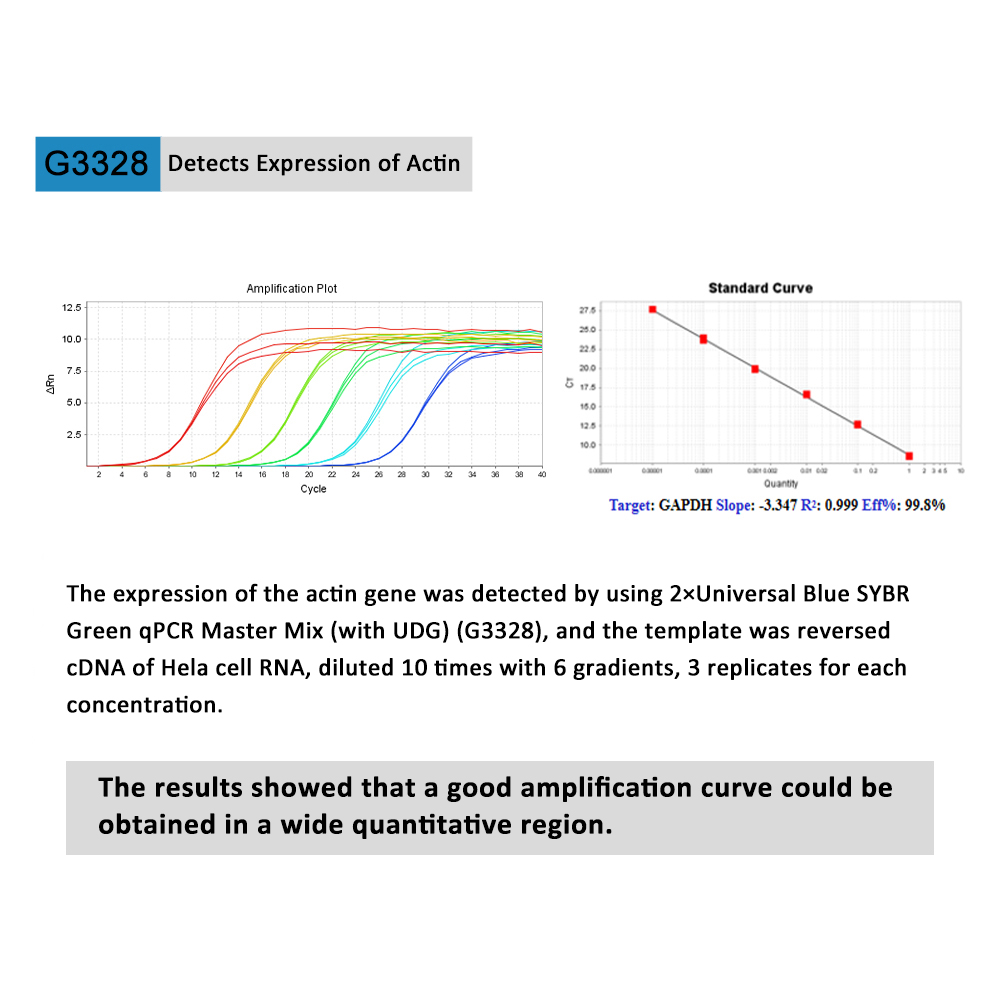
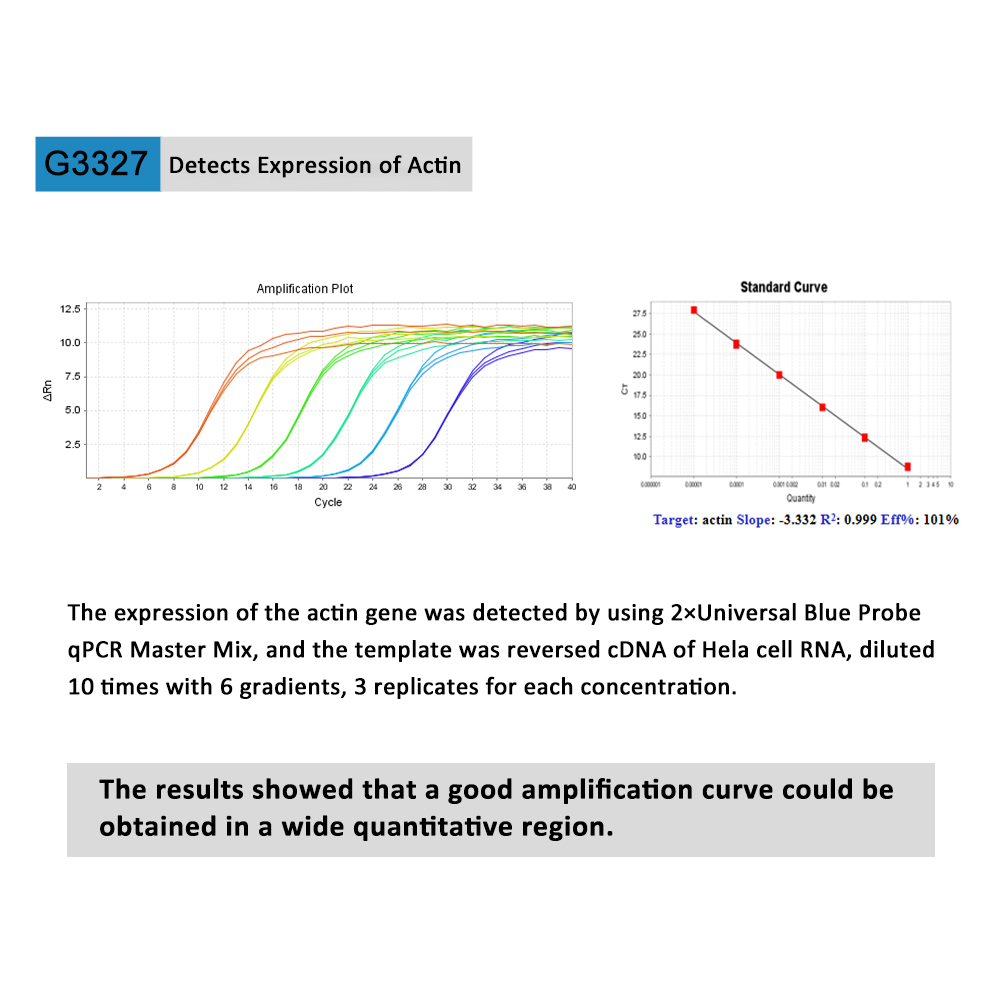
This product is a special 2× premixed solution for qPCR reaction using SYBR Green I chimeric fluorescence method. It contains all qPCR components except primers and DNA templates, which can reduce the operation steps, shorten the sample addition time and decrease the probability of contamination. The core component is the genetically engineered heat-activated Taq DNA Polymerase, which effectively blocks DNA Polymerase activity and prevents nonspecific amplification at low temperatures by efficiently binding monoclonal antibodies to the Taq DNA Polymerase. The heat-activated Taq DNA Polymerase has many advantages such as high specificity and detection sensitivity. With the reaction buffer optimized for qPCR, it is very suitable for high-specificity and high-sensitivity qPCR reactions. This product is a 2× premixed reagent containing the optimal concentration of SYBR Green I for qPCR reaction, which can obtain a good standard curve in a wide quantitative area. The quantification of target genes is accurately, reproducible and reliable.
Storage and Handling Conditions
Transport with wet ice. Store at -20°C without light, valid for 12 months. Avoid freeze/thaw cycle. After thawing, it can be stably stored at 4℃ for one month without light.
Component
| Component | G3322-01 | G3322-05 | G3322-15 |
| 2×SYBR Green qPCR Master Mix (High ROX) | 1 mL | 5×1 mL | 15×1 mL |
| Manual | One copy | ||
Assay Protocol / Procedures
Preparation before experiment
1. Real Time PCR amplification apparatus;
2. Special reaction tube or reaction plate for experiment;
3. PCR primers (reference primer design principles);
4. Micropipette and pipette head (autoclaving);
Procedures
1. Recommend the qPCR reaction system:
| Component | 20 μL rxn | 50 μL rxn | Final Concentration |
| 2×SYBR Green qPCR Master Mix (High ROX) | 10 μL | 25 μL | 1× |
| Forward Primer (10 μM)a | 0.4 μL | 1 μL | 0.2 μM |
| Reverse Primer (10 μM)a | 0.4 μL | 1 μL | 0.2 μM |
| Templateb | Variable | Variable | as required |
| Nuclease-Free Water | Add to 20 μL | Add to 50 μL |
a. Usually, a good amplification effect can be obtained with the final concentration of 0.2 μM. When the reaction performance is poor, the primer concentration can be adjusted in the range of 0.2-1.0 μM.
b. The amount of template addition varies with the copy number of the target gene in the template solution, and the appropriate amount of template addition is discussed by gradient dilution. The best addition amount of template DNA in the 20 μL reaction system was less than 100 ng. When the cDNA (RT reaction solution) of RT-PCR reaction was used as template, the addition amount should not exceed 10% of the total volume of PCR reaction solution.
2. PCR reaction procedure (can be adjusted according to the instruments):
*: If amplification specificity needs to be improved, two-step procedure or annealing temperature can be used; To improve the amplification efficiency, a three-step procedure or extension time can be used.
a: For fluorescence signal collection, please set the experimental procedure according to the instruction manual of the instrument.
Note
1. Mixed gently upside down before use. Do not swirl and shake to avoid bubbles.
2. Reagents should be placed on ice when preparing reaction solution.
3. The product contains fluorescent dye SYBR Green, so strong light should be avoided when preparing PCR reaction solution.
4. Please using new disposable head for the preparation and packaging of the reaction solution to avoid contamination between samples.
5. Avoid repeated freeze-thaw Master Mix and try to use it within one month after thawing.
Compatibility
Primer Design Principles
1. The length of amplification product is recommended to be between 80-300 bp;
2. Primer length: 18-25 bp;
3. The content of base G+C in primers should be between 40%-60%;
4. The Tm value difference between forward primers and reverse primers is less than 2℃, and the Tm value between 58-62℃ is the best;
5. Randomness of base distribution;
6. Primers had better not contain self-complementary sequences, otherwise they will form a secondary hairpin structure;
7. There should be no more than 4 complementary or homologous bases between two primers, otherwise primer dimer will be formed, especially complementary overlap at the 3′ end;
8. The 3′ terminal base of the primer is suggested to be G or C;
9. No other non-specific products were found in NCBI comparison results.
Trouble-Shooting
| Problem description | Possible reasons | Solutions |
| At the end of the reaction, no amplification curve appeared or CT value appeared too late | The template concentration is too low | Repeat the experiment to reduce the template dilution multiple, and start from the highest concentration when the sample concentration is unknown |
| Template degradation | The template was prepared again and the experiment was repeated | |
| There are PCR inhibitors in the system | Generally, the template is carried in, the dilution ratio of the template is increased or the template with high purity is reprepared and repeated | |
| Primers may degrade | Primers that have not been used for a long time should first be tested for integrity by PAGE electrophoresis to rule out the possibility of degradation | |
| Low amplification efficiency | Increase the primer concentration, try a three-step amplification procedure, or redesign the primer | |
| The amplification product is too long | The amplification product length was controlled in the range of 80-300 bp | |
| The blank control shows the signal | Reaction system pollution | Firstly, the blank control water should be replaced. If the same situation still occurs, the primers, aspirators and PCR tubes should be replaced or a new Master Mix should be started. The reaction system is prepared in a super clean table to reduce aerosol pollution |
| Non-specific amplification such as primer dimers appears | Generally, it is normal for the amplification products to appear in blank control after 35 cycles, which should be analyzed with the melting curve.
Redesign primer, adjust primer concentration or optimize PCR reaction procedure |
|
| The melting curve has multiple peaks | Primer design is poor | The new primer was re-designed according to the primer design principles |
| Primer concentration is too high | Reduce primer concentration appropriately | |
| There is genomic contamination in cDNA template | The extracted RNA solution is digested using DNA enzymes, such as dsDNase, to remove genomic contamination, or to design transintron primers | |
| Poor reproducibility of experiments | The error of adding sample is large | The use of accurate pipette, with high quality suction head accurate pipette;
High dilution template, adding large volume template to reduce sampling error; The reaction volume of qPCR was enlarged |
| The template concentration is too low | Repeat the experiment to reduce the dilution times of the template | |
| Temperature deviation at different locations of the qPCR instrument | Calibrate the qPCR instrument regularly | |
| The amplification curve is not smooth | Fluorescence signal is too weak, produced after system correction | Ensure that the dyes premixed in the Master Mix are not degraded;
Replace fluorescent signal to collect better qPCR consumables |
| Amplification curve breaks or slips | The template concentration was higher and the baseline endpoint value was greater than the CT value | The baseline endpoint (Ct value -3) was reduced and the data were reanalyzed |
| Amplification curves of individual Wells suddenly dropped sharply | There are bubbles in the reaction tube | Ensure that MIX is completely dissolved, and do not swirl and oscillate evenly;
After the sample is added, the bubbles are removed by centrifugation with light elastic. The pre-denaturation time was extended to 10 min to remove the bubbles |
For Research Use Only!
Ver. No.: V1.0-202102
|
Cat.No.
|
Product Name
|
Spec.
|
Operation
|
|---|
|
G3001-500ML
|
50×TAE Electrophoresis Buffer Solution
|
500 mL
|
|
|
G3002-250ML
|
10×TBE Electrophoresis Buffer Solution
|
250 mL
|
|
|
G3003-250ML
|
Servicebio® 10×TE(pH 8.0)
|
250 mL
|
|
|
G3004-100ML
|
DEPC Water
|
100 mL
|
|
|
G3013-100ML
|
RNA Extraction Solution
|
100 mL
|
|
|
G3320-05
|
2 × SYBR Green qPCR Master Mix (None ROX)
|
5 mL
|
|
|
G3321-05
|
2×SYBR Green qPCR Master Mix (Low ROX)
|
5 mL
|
|
|
G3322-05
|
2 × SYBR Green qPCR Master Mix (High ROX)
|
5 mL
|
|
|
G3323-05
|
2 × Fast SYBR Green qPCR Master Mix (None ROX)
|
5 mL
|
|
|
G3324-05
|
2 × Fast SYBR Green qPCR Master Mix (Low ROX)
|
5 mL
|
|
|
G3325-05
|
2 × Fast SYBR Green qPCR Master Mix (High ROX)
|
5 mL
|
|
|
G3330-100
|
SweScript RT I First Strand cDNA Synthesis Kit
|
100 rxns
|
|
|
G3340-100
|
T4 DNA Ligase (5 U/μL )
|
500 U
|
|
|
G5042-1G
|
Isopropyl β-D-1-thiogalactopyranoside (IPTG)
|
1 g
|
|
|
G5056-5G
|
Agarose(Electrophoresis-Grade, Low EEO)
|
5 g
|