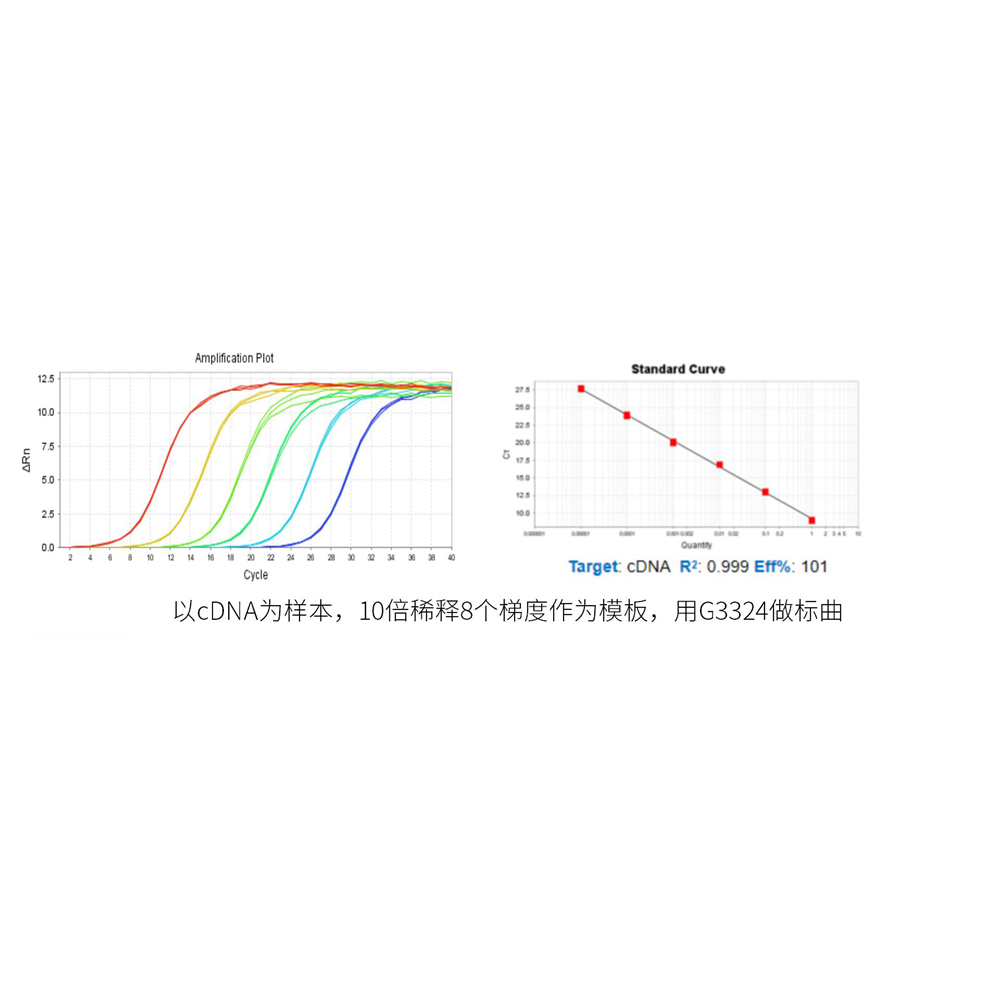
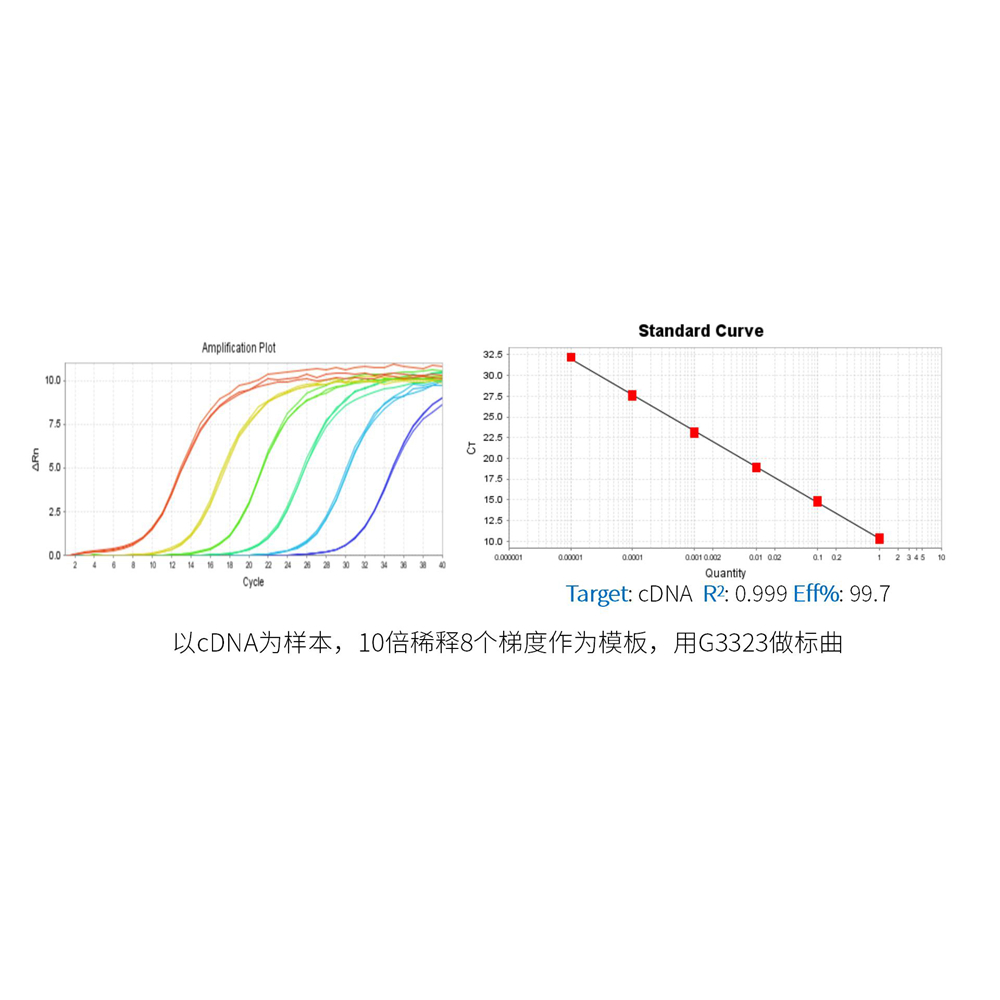
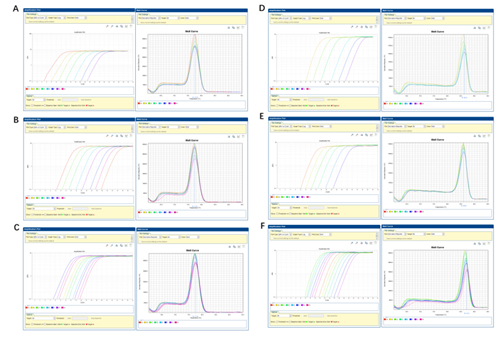
Description
Product Information
| Product Name | Cat. No. | Spec. |
| 2×Universal Blue SYBR Green qPCR Master Mix (with UDG) | G3328-01 | 1 mL |
| G3328-05 | 5×1 mL | |
| G3328-15 | 15×1 mL |
Product Description
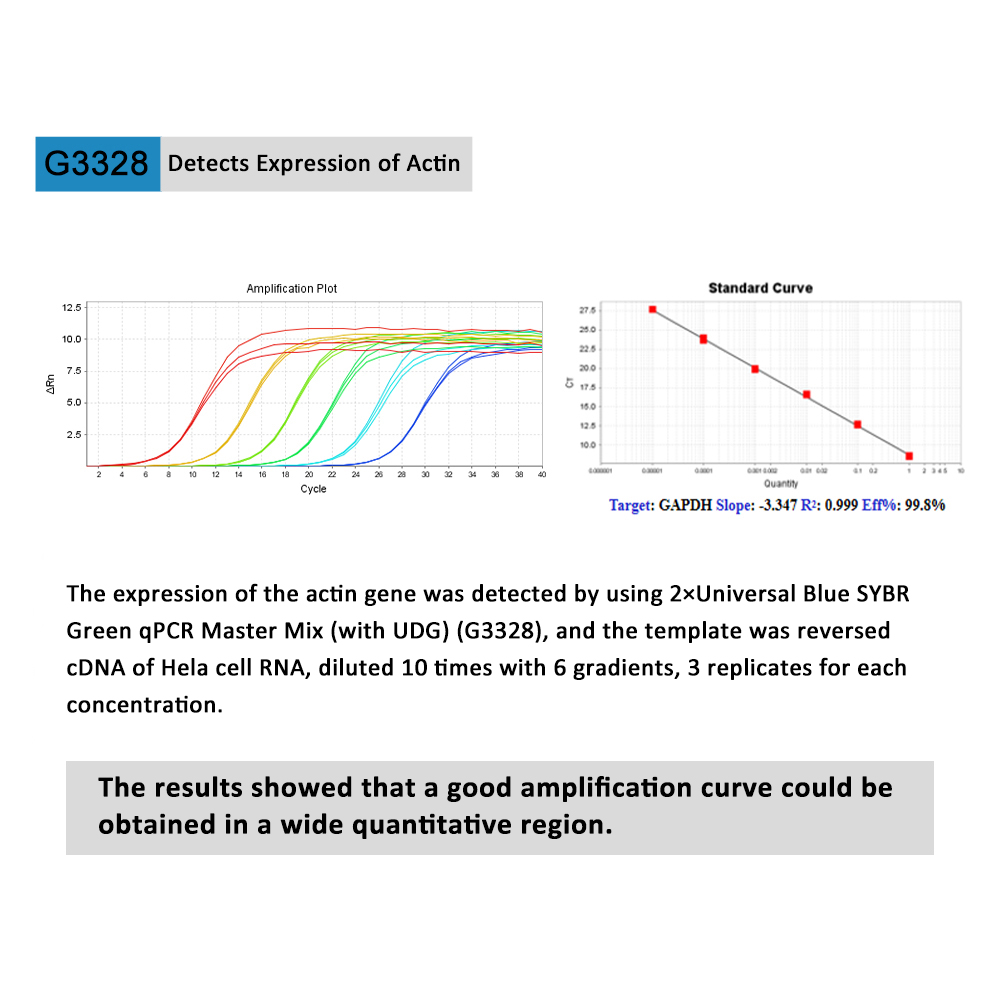
The 2×Universal Blue SYBR Green qPCR Master Mix (with UDG) is a pre-formulated, optimized, universal 2X master mix for real-time PCR workflows with built-in visual dye–based indicators for accurate reaction set up. Coupled with user-supplied primer sets and template, the Master Mix is designed to amplifiy targets for accurate gene expression analysis.
it contains Taq DNA polymerase, which is tightly controlled through an antibody-mediated hot-start mechanism, helping prevent undesirable early activity of the polymerase at low temperatures that can lead to nonspecific amplification, and formulated with UNG/dUTP to prevent contamination of carryover PCR products. Meanwhile,it contains an inert blue dye that does not interfere with real-time PCR but provides increased visibility in the tube or well. A separate sample buffer is provided that contains an inert yellow dye. The optional use of this yellow sample buffer results in a reaction mixture that changes from blue to green upon addition of the sample, helping one avoid pipetting mistakes. The yellow sample buffer aids in reaction setup, but is not required to obtain superior results with the Master Mix.Those two blue and yellow dyes do not spectrally overlap fluorophores commonly used for qPCR and will not interfere with real-time detection.
Storage and Shipping Conditions
Ship with wet ice; store at -20°C away form light, valid for 12 months.
Product Contents
| Component Number | Component | G3328-01 | G3328-05 | G3328-15 |
| G3328-1 | 2×Universal Blue SYBR Green qPCR Master Mix (with UDG) | 1 mL | 5×1 mL | 15×1 mL |
| G3328-2 | 40×Yellow Template Dilution Buffer | 1 mL | 1 mL | 1 mL |
| Specification | 1 copy | |||
Assay Protocol
Prepare the PCR reactions
Note: The Yellow Sample Buffer is optional for the real-time PCR.
The Yellow Sample Buffer is supplied at a 40X concentration. It is added to the DNA template. The concentration of Yellow Sample Buffer in the final PCR must be 1X. It is recommended that the DNA template is 10–20% of the volume of the final PCR.
1. (Optional) Add the Yellow Sample Buffer (40X) to the amount of DNA that is used in the PCR.
Taking the 20-μL qPCR reaction system as an example, according to the dilution template amount added into the 20μL qPCR reaction solution, the corresponding dilution method of the original template can be referred to the following table (taking the original template diluted to 100μL as an example):
| Add the diluted template to the 20 μL qPCR reaction solution (μL) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Add the 40x YELLOW Template Dilution Buffer to the 100 μL Template Dilution System (μL) | 50 | 25 | 16.7 | 12.5 | 10 | 8.4 | 7.2 | 6.3 |
| Add the 100μL Template Dilution System to the primary template (μL) | x | x | x | x | x | x | x | x |
|
Volume of adding Nuclease – Free Water (μL) |
50-x | 75-x | 83.3-x | 87.5-x | 90-x | 91.6-x | 92.8-x | 93.7-x |
For example: 2μL diluted template should be added to 20 μL qPCR reaction system. Taking the 100μL Template Dilution system as an example. If the original template volume is 20μL, 25μL 40x Yellow Template Dilution Buffer is added, and then Nuclease-free Water 55μL is added to the total volume of 100μL. The 40x YELLOW Template Dilution Buffer is now 10x. Add 2μL of the diluted template into a 20-μL qPCR reaction system, and the final concentration of 40× YELLOW Template Dilution Buffer is 1x.
Note: If the concentration of the template is too low, do not need to dilute, or do not want to use diluted template with 40x YELLOW Template Dilution Buffer, you can omit this step.
2. (Optional) Vortex, then centrifuge the DNA and Yellow Sample Buffer.
3. Combine the master mix, the primers, and nuclease-free water with the DNA and Yellow Sample Buffer according to the following tables.
| Component | 20 μL rxn | 50 μL rxn | Final Concentration |
| 2×Universal Blue SYBR Green qPCR Master Mix (with UDG) | 10 μL | 25 μL | 1× |
| Forward Primer (10 μM)a | 0.4 μL | 1 μL | 0.2 μM |
| Reverse Primer (10 μM)a | 0.4 μL | 1 μL | 0.2 μM |
| Templateb | Variable | Variable | as required |
| Nuclease-free Water | Add to 20 μL | Add to 50 μL |
a: Usually, a good amplification effect can be obtained with the final concentration of 0.2 μM. When the reaction performance is poor, the primer concentration can be adjusted in the range of 0.2-1.0 μM.
b: The amount of template added varies depending on the number of copies of the target gene, and the appropriate amount of template addition is studied by gradient dilution. The best addition amount of template DNA in the 20 μL reaction system was less than 100 ng. When the cDNA (RT reaction solution) of RT-PCR reaction was used as template, the addition amount should not exceed 10% of the final qPCR volume.
Note: If the Yellow Sample Buffer is not used, add nuclease-free water to achieve the total PCR volume.
4. Mix the components thoroughly, then centrifuge briefly to collect the contents at the bottom of the tube.
5. Transfer the appropriate volume of each reaction to each well of an optical plate.
6. Seal the plate with an optical adhesive cover, then centrifuge briefly to collect the contents at the bottom of each well and eliminate any air bubbles.
| A. Two-step method | B. The three-step method | ||||||||
| stage | step | Cycle number | Temp
erature |
time | stage | step | Cycle number | Temp
erature |
time |
| Stage 1 | UDG incubation | 1 | 50℃ | 2
min |
Stage 1 | UDG incubation | 1 | 50℃ | 2
min |
| Stage 2 | predegeneration | 1 | 95℃ | 30 sec | Stage 2 | Predeg
eneration |
1 | 95℃ | 30 sec |
| Stage 3 | denaturation | 40 | 95℃ | 15 sec | Stage 3 | denaturation | 40 | 95℃ | 15 sec |
| Annealing
/extension |
60℃ | 30 seca | Annealing | 55-65℃ | 10 sec | ||||
| Extension | 72℃ | 30 seca | |||||||
| Stage 4 | Melting curve | 1 | Instrument
default setting |
Stage 4 | Melting curve | 1 | Instrument
default setting |
||
7. Program the thermal cycler according to the recommendations below, place the samples in the cycler and start the program.
a: If amplification specificity needs to be improved, two-step procedure or annealing temperature can be used; To improve the amplification efficiency, a three-step procedure or extension time can be used.
Note
1. If pipetting tracking is not required, dilute template with the 40×Yellow Template Dilution Buffer can be omitted.
2. Please Mix the reaction mix gently by inversion, then centrifuge briefly before aliquoting to the reaction plate to avoid bubbles.
3. Reagents should be placed on ice when preparing reaction mixes..
4. Strong light should be avoided when preparing PCR reaction mixes due to fluorescent dye SYBR Green.
5. New disposable tips should be used for preparation of reaction mixes to avoid cross contamination
6. Avoid freeze-thawing cycles of the Master Mix, and try to use it up within a month after thawing.
Compatible Instruments
ABI: 5700, 7000, 7300, 7700, 7900, 7900HT, 7900HT Fast, StepOne™, StepOne Plus™, 7500/7500 Fast, ViiA 7™, QuantStudio™ Series, PikoRealTM Cycler.
Stratagene: Mx3000P®, 3005P™,
Bio-Rad: CFX96™, CFX384™, iCycler iQ™, iQ5™, MyiQ™, MiniOpticon™, Opticon®, Opticon 2, Chromo4™;
Eppendorf: Realplex 2s, Mastercycler® ep, realplex;
IIIumina: Eco QPCR
Cepheid: SmartCycler®;
Qiagen Corbett: Rotor-Gene® series;
Roche: LightCycler™ Series;
Takara: Thermal Cycler Dice series;
Analytikjena: qTOWER series;
Hangzhou Bori: LineGene department;
Primer Design Principles
1. The length of the amplified product is recommended to be between 80-300 bp;
2. Primer length: 18-25 bp;
3. Keep the GC content in the 40%-60% range.
4. Optimal melting temperature (Tm): 60°C. Differences in Tm of the two primers should not exceed 2°C;
5. Avoid runs of identical nucleotides. If repeats are present, there must be fewer than four consecutive G residues;
6. Avoid self-complementarities in a primer, complementarities between the primers and direct repeats in a primer to prevent hairpin formation and primer dimerization.
7. Make sure the last five nucleotides at the 3′ end contain no more than two G and/or C bases.
8. No non-specific products appear on NCBI blast.
Trouble-Shooting
| Problem | Possible Causes | Solutions |
| No amplification curve appeared or CT value appeared too late | Annealing temperature is not optimal. | Optimize the annealing temperature in 3°C increments. |
| Degradation of primers. | Check PCR primers for possible degradation on polyacrylamide gel. | |
| PCR inhibitors present in the reaction mixture. | Re-purify your template DNA. | |
| Primer design is suboptimal | Verify your primer design, use reputable primer design programs or validated pre-designed primers. | |
| RT-qPCR: inhibition by excess volume of the RT reaction. | Volume of RT reaction product added to qPCR reaction should not exceed 10% of the total qPCR reaction volume. | |
| Pipetting error or missing reagent. | Repeat the PCR reaction; check the concentrations of template and primers; ensure proper storage conditions of all reagents. Make new serial dilutions of template DNA or RNA. | |
| Amplification signal in non-template control | DNA contamination of reagents. | · Follow general guidelines to avoid carry over contamination; Discard reagents and repeat with new reagents. |
| Primer-dimers. | Use melting curve analysis to identify primer-dimers by the lower melting temperature compared to amplicon | |
| RT-qPCR: RNA contaminated with genomic DNA | Design primers on intron/exon boundaries, treat RNA sample with DNaseI,RNA free prior to reverse transcription.
|
|
| The melting curve has multiple peaks | Primer design is suboptimal | Verify your primer design, use reputable primer design programs or validated pre-designed primers. |
| Primer concentration is too high | Reduce primer concentration appropriately | |
| Genomic contamination in cDNA template | The extracted RNA solution is digested using DNA enzymes, such as dsDNase, to remove genomic contamination, or to design transintron primers | |
| Poor reproducibility of experiments | The error of adding sample is large | The use of accurate pipette, with high quality suction head accurate pipette;
High dilution template, adding large volume template to reduce sampling error; The reaction volume of qPCR was enlarged |
| The template concentration is too low | Repeat the experiment to reduce the dilution times of the template | |
| Temperature deviation at different locations of the qPCR instrument | Calibrate the qPCR instrument regularly | |
| The amplification curve is not smooth | Fluorescence signal is too weak, produced after system correction | Ensure that the dyes premixed in the Master Mix are not degraded;
Replace fluorescent signal to collect better qPCR consumables |
| Amplification curve breaks or slips | The template concentration was higher and the baseline endpoint value was greater than the CT value | The baseline endpoint (Ct value -3) was reduced and the data were reanalyzed |
| Amplification curves of individual Wells suddenly dropped sharply | There are bubbles in the reaction tube | Ensure that MIX is completely dissolved, and do not swirl and oscillate evenly;
After the sample is added, the bubbles are removed by centrifugation with light elastic. The pre-denaturation time was extended to 10 min to remove the bubbles |
For Research Use Only!
|
Cat.No.
|
Product Name
|
Spec.
|
Operation
|
|---|
|
G3326-01
|
2×Universal Blue SYBR Green qPCR Master Mix With 40×Yellow Template Dilution Buffer
|
1 mL
|
|
|
G3326-05
|
2×Universal Blue SYBR Green qPCR Master Mix With 40×Yellow Template Dilution Buffer
|
5×1 mL
|
|
|
G3326-15
|
2×Universal Blue SYBR Green qPCR Master Mix With 40×Yellow Template Dilution Buffer
|
15×1 mL
|
|
|
G3327-01
|
2×Universal Blue Probe qPCR Master Mix
|
1 mL
|
|
|
G3327-05
|
2×Universal Blue Probe qPCR Master Mix
|
5×1 mL
|
|
|
G3327-15
|
2×Universal Blue Probe qPCR Master Mix
|
15×1 mL
|
|
|
G3328-01
|
2×Universal Blue SYBR Green qPCR Master Mix (with UDG)
|
1 mL
|
|
|
G3328-05
|
2×Universal Blue SYBR Green qPCR Master Mix (with UDG)
|
5×1 mL
|
|
|
G3328-15
|
2×Universal Blue SYBR Green qPCR Master Mix (with UDG)
|
15×1 mL
|