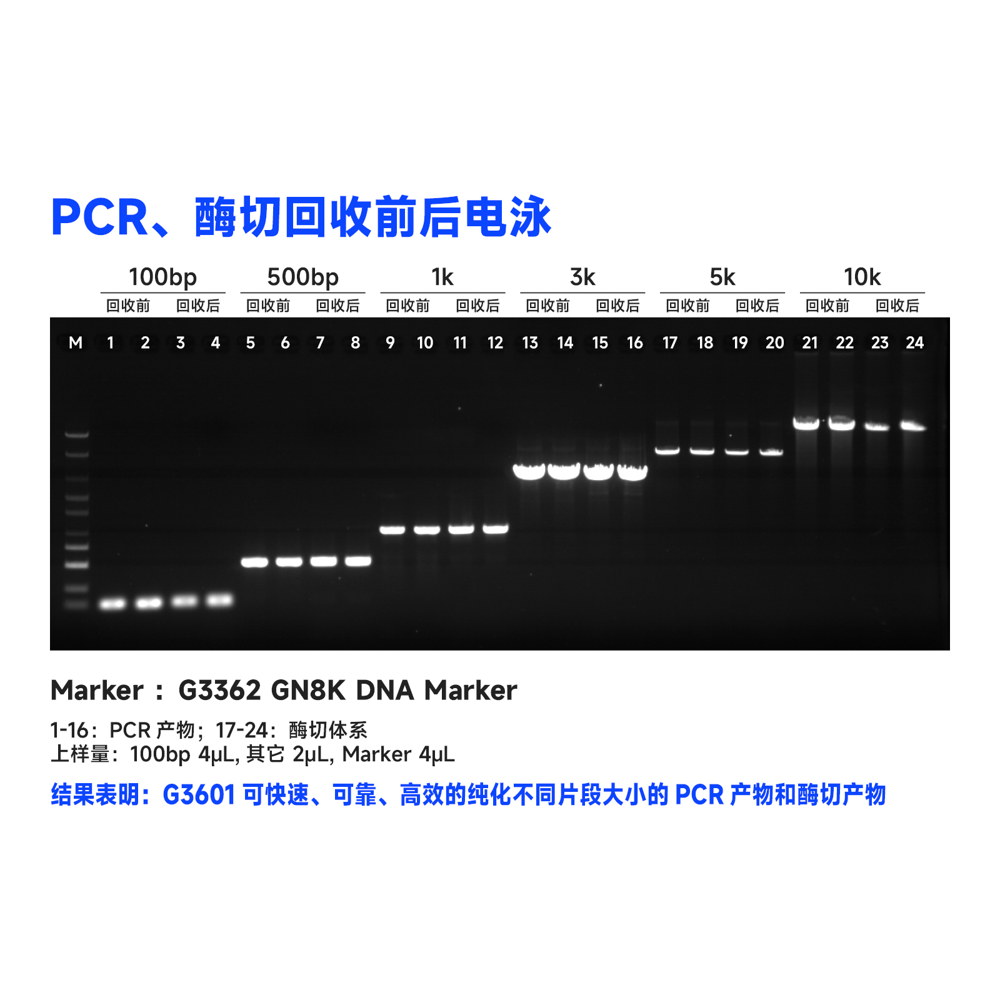
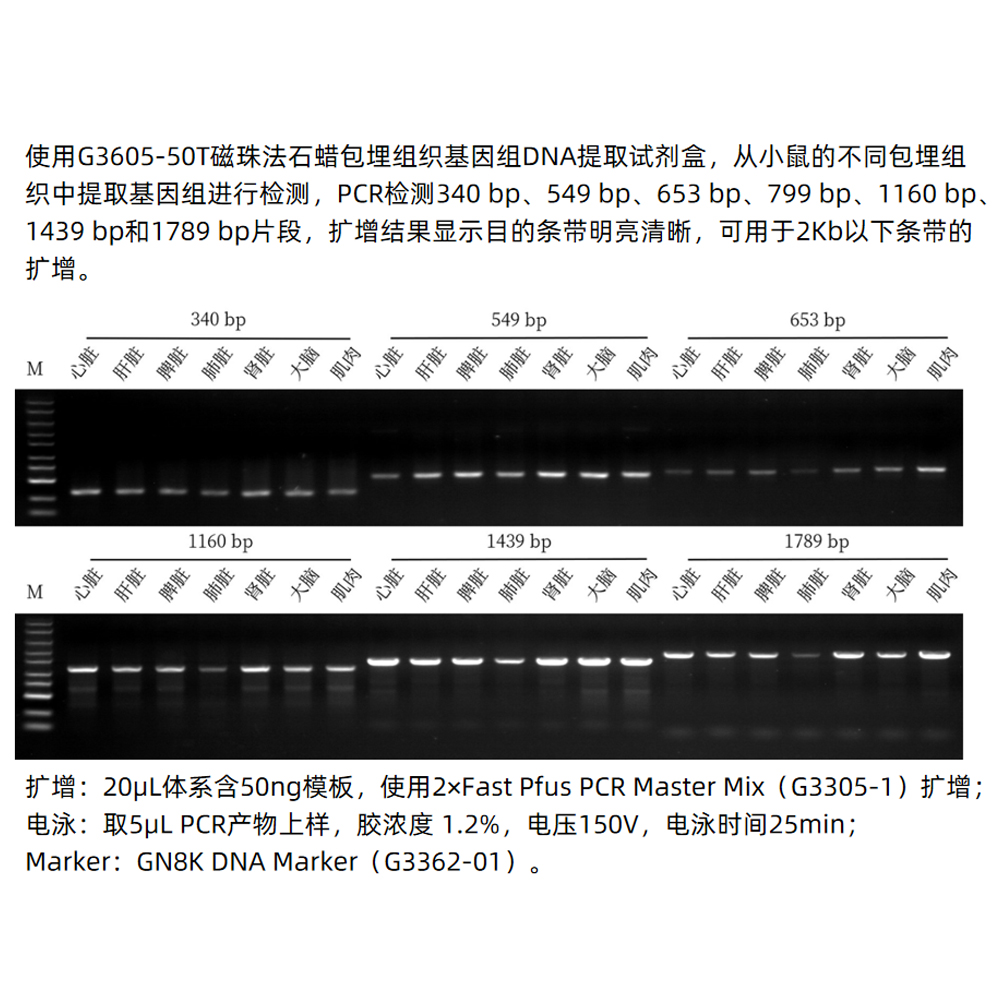
Description
The provided instructions appear to be for a “Magnetic Bead-based Bacterial Genomic DNA Extraction Kit.” Here is the translation of these instructions into English:
Product Information
- Product Name: Magnetic Bead-based Bacterial Genomic DNA Extraction Kit
- Product Number: G3603-50T
- Specifications: 50 tests
Product Overview
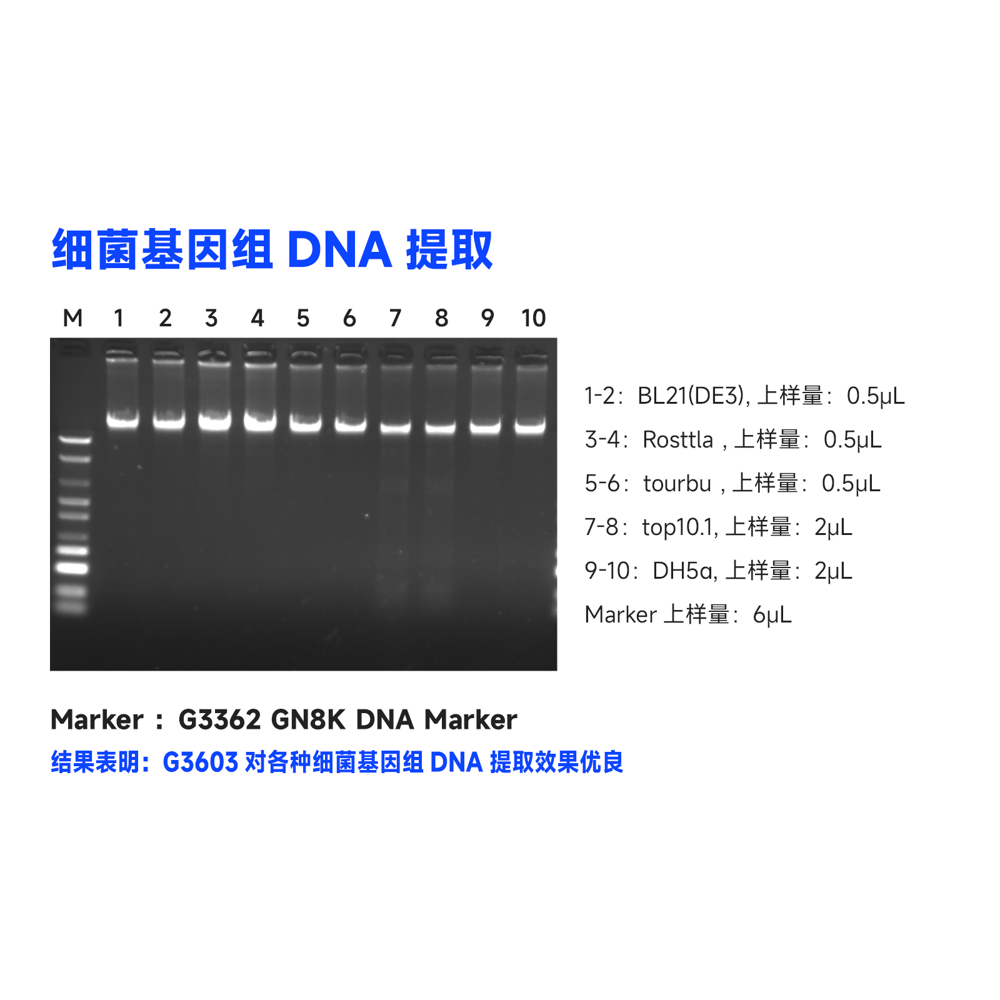
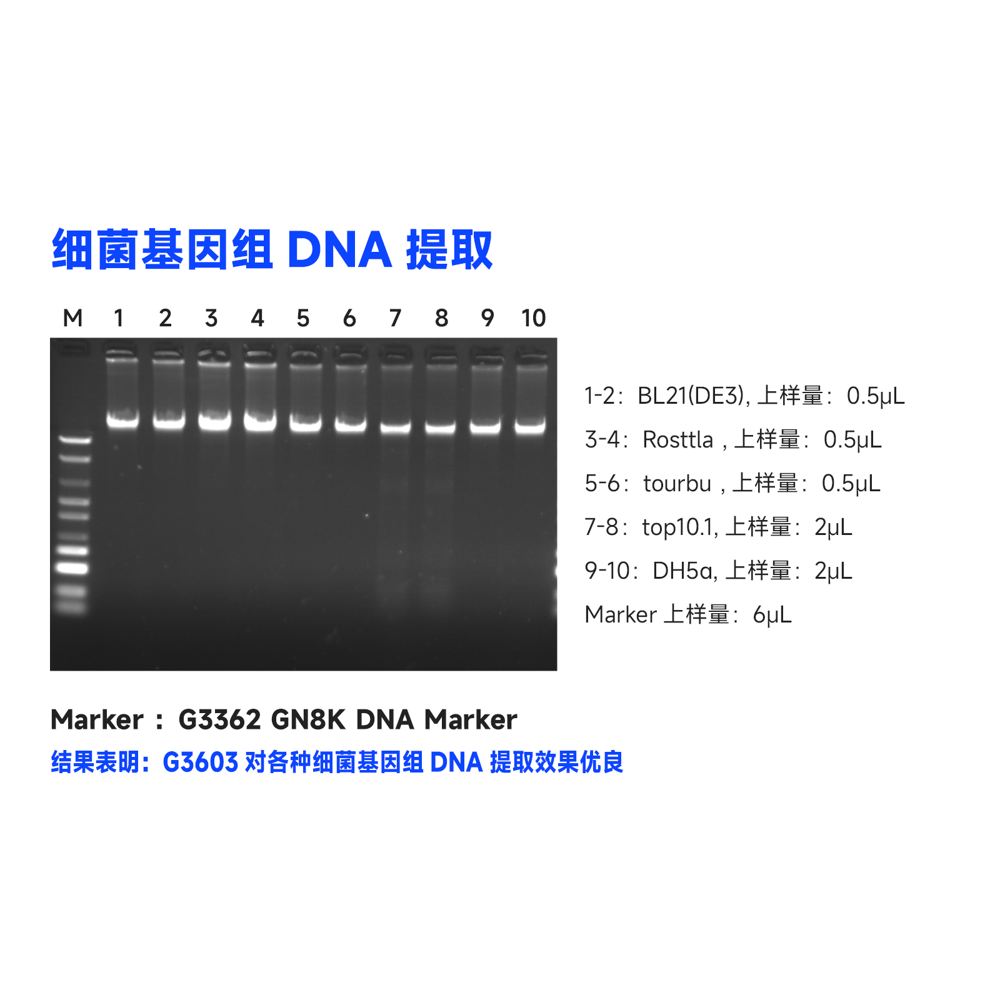
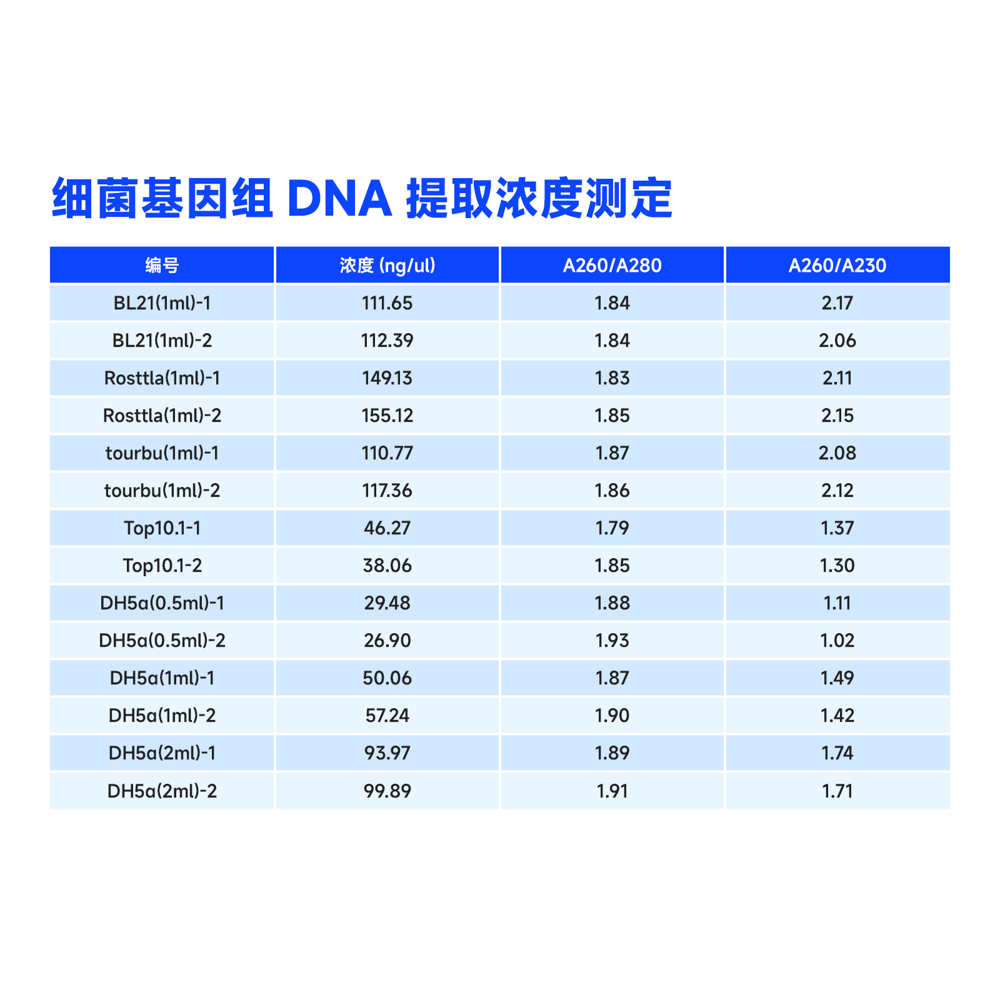
This kit uses specially optimized superparamagnetic beads in conjunction with specific lysis and binding solutions to selectively adsorb bacterial genomic DNA. Impurities on the surface of the beads are removed with a washing solution, and the genomic DNA bound to the beads is eluted using an elution solution, resulting in high-quality genomic DNA. This product can rapidly purify 10-50 μg of high-purity, high-integrity genomic DNA from 1-5 mL of fresh bacterial culture. The obtained genomic DNA can be directly used in subsequent molecular biology experiments such as PCR amplification, qPCR, enzyme digestion reactions, Southern blotting, library construction, and more.
Storage and Transportation
- Lysozyme, Proteinase K, and RNase A are shipped on wet ice and should be stored at -20°C.
- Other reagents are shipped and stored at room temperature.
- Shelf life is 12 months.
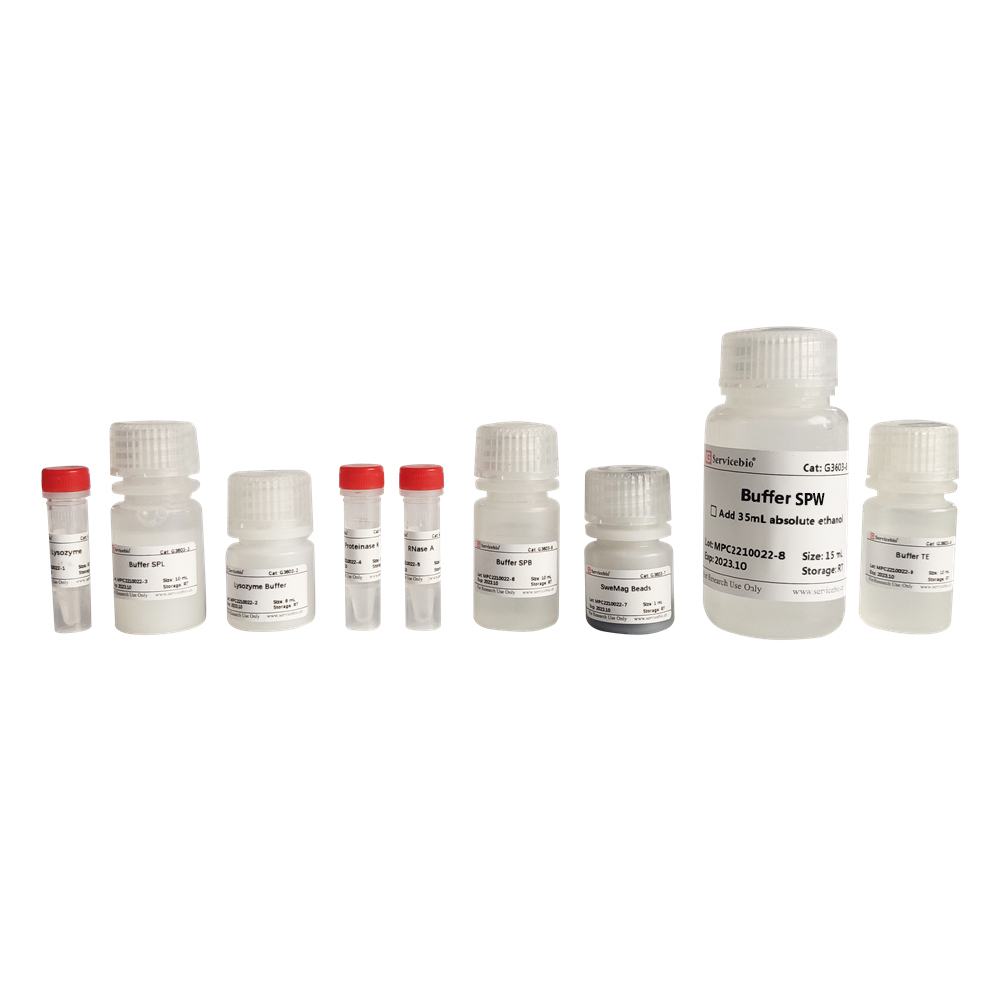
Components
| Component Number | Component | Volume |
|---|---|---|
| G3603-1 | Lysozyme | 500 µL |
| G3603-2 | Lysozyme Buffer | 6 mL |
| G3603-3 | Buffer SPL | 10 mL |
| G3603-4 | Proteinase K | 1 mL |
| G3603-5 | RNase A | 500 µL |
| G3603-6 | Buffer SPB | 10 mL |
| G3603-7 | SweMag Beads | 1 mL |
| G3603-8 | Buffer SPW | 15 mL |
| G3603-9 | Buffer TE | 10 mL |
| Instruction Manual | 1 copy |
Preparation Before Use
- If Buffer SPL has precipitation, heat it at 65°C until the precipitation disappears, mix well before use.
- Before use, add the specified amount of ethanol to Buffer SPW (as indicated on the bottle).
- Prepare a magnetic rack and 1.5 mL sterile centrifuge tubes.
Experimental Procedure
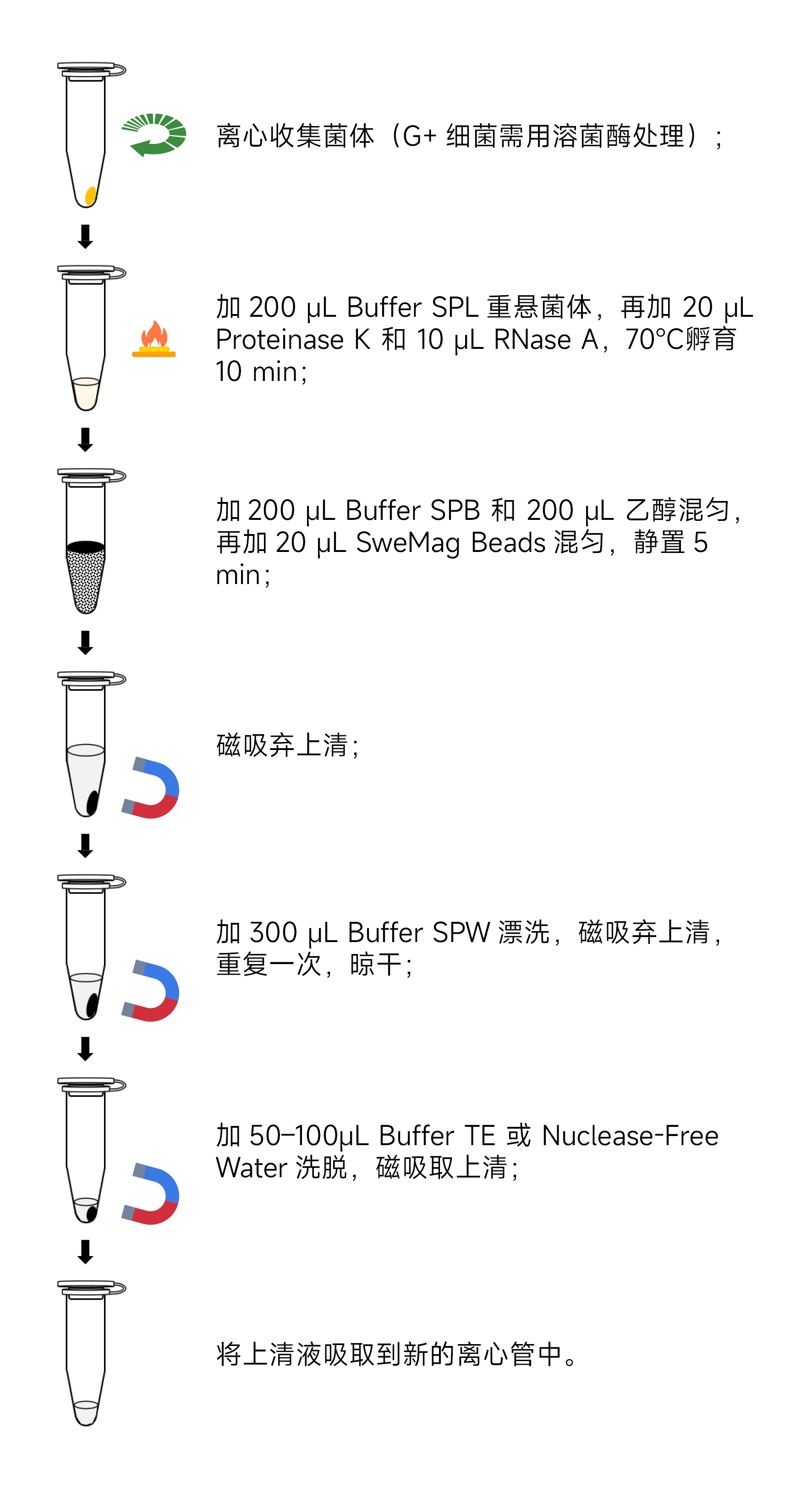
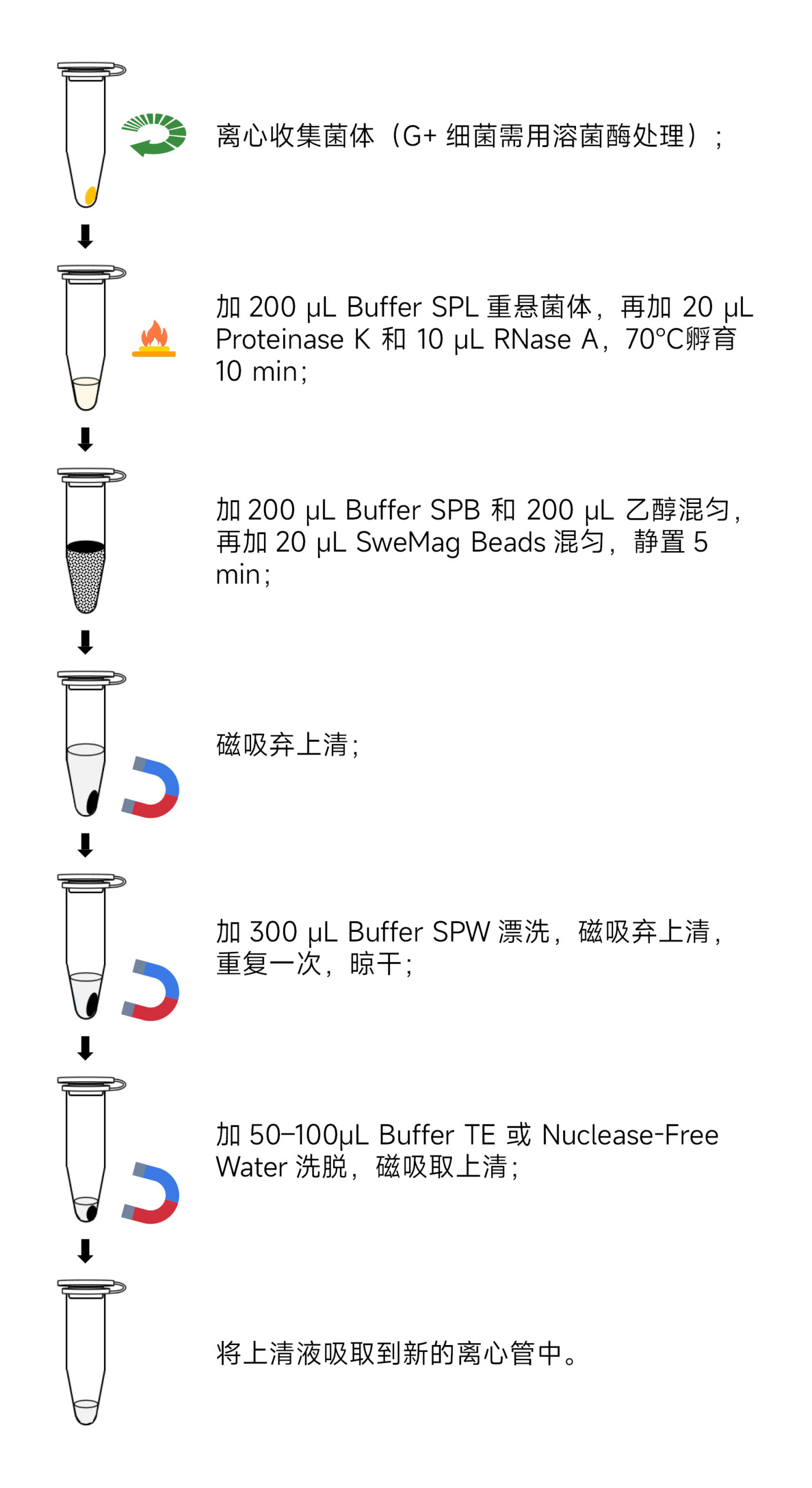
- Sample Preparation:
- For bacterial cultures, take 1-5 mL of overnight culture, centrifuge at 12,000 rpm for 1 min, collect the pellet in a 1.5 mL centrifuge tube, and discard the supernatant.
- For Gram-positive bacteria that are difficult to lyse, add lysozyme for cell wall digestion (see detailed instructions).
- Add 200 μL of Buffer SPL to the tube from step 1, disperse the bacterial pellet thoroughly using a pipette or vortex.
- Add 20 μL Proteinase K and 10 μL RNase A, incubate at 70°C for 10 min until the solution becomes transparent. If not transparent, extend the reaction time to 30 min, pipetting up and down every 5 min.
- Add 200 μL of Buffer SPB and 200 μL of anhydrous ethanol, invert the tube several times to mix, and then add 20 μL of SweMag Beads (vortex well before use). Invert the tube several times to ensure even dispersion of the beads.
- Incubate at room temperature for 5 minutes, mixing by pipetting or vortexing 2-3 times during incubation to maintain even bead dispersion.
- Place the tube on a magnetic rack for 30 seconds until the beads are completely adsorbed. Then, gently invert the tube several times along with the magnetic rack to wash off any residual beads attached to the tube wall. Discard the supernatant (do not aspirate the beads to avoid affecting extraction efficiency).
- Add 300 μL of Buffer SPW to the tube, remove it from the magnetic rack, and gently pipette to ensure even bead dispersion. Place the tube back on the magnetic rack for 30 seconds. Again, gently invert the tube several times along with the magnetic rack to wash off any residual beads and salt. Discard the supernatant (be sure to remove all liquid from the tube to avoid affecting extraction efficiency).
- Repeat step 7.
- Leave the tube open at room temperature for 5-10 minutes or at 65°C for 3-5 minutes to allow ethanol to completely evaporate (avoid over-drying the beads to prevent loss of genomic DNA yield).
- Remove from the magnetic rack, add 50-100 μL of Buffer TE or nuclease-free water to the tube, pipette gently to ensure even bead dispersion, and leave at room temperature for 3 minutes.
- Place the tube back on the magnetic rack until the beads are completely adsorbed, then transfer the supernatant to a new centrifuge tube to obtain high-purity genomic DNA.
Precautions
- Before operation, please read the product manual carefully.
- The yield and integrity of extracted DNA depend on the type of sample, fixation time, fixation conditions, and sample storage time. It is best to fix samples within 8-24 hours. If the sample fixation time exceeds 24 hours or the storage time exceeds 1 year, it may result in excessive DNA fragmentation and the inability to amplify the desired bands.
- Samples should be completely dehydrated before embedding to prevent residual formalin from affecting subsequent experiments.
- When sampling embedded tissues, try to remove excess paraffin and do not exceed 30 mg of the sample. Excessive samples can lead to incomplete lysis, affecting DNA yield.
- Magnetic beads can precipitate, so shake or vortex well before use. Magnetic bead suspensions should be stored without freezing.
- Before eluting genomic DNA, ensure that ethanol is completely evaporated to avoid residual ethanol affecting downstream experiments. Do not overdry the beads to prevent a decrease in genomic DNA yield.
- When using the extracted genomic DNA as a template for PCR amplification, it is recommended to perform a template concentration gradient test to select the optimal template concentration for amplification.
- For your safety and health, please wear a lab coat and disposable gloves while operating.
This product is for research use only and is not intended for clinical diagnosis.