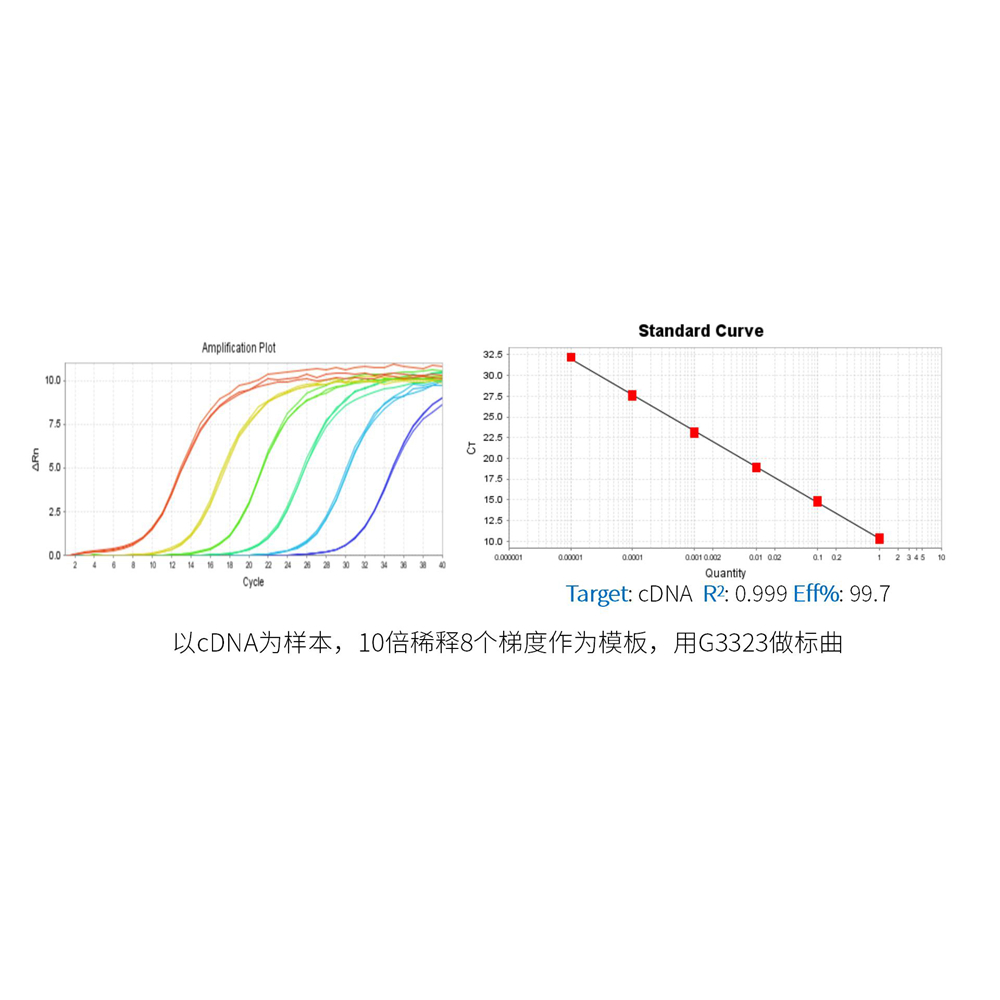
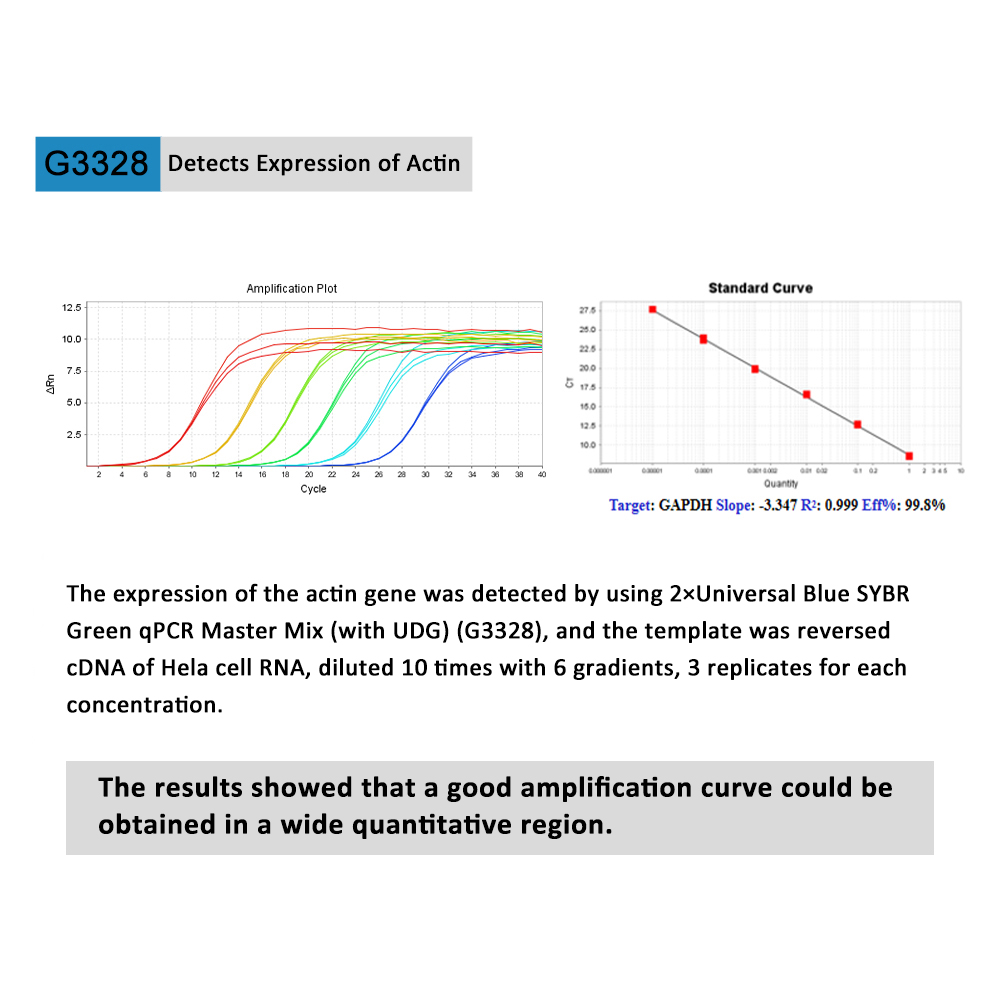
Description
Product Information
| Product Name | Cat. No. | Spec. |
| 2×Universal Blue Probe qPCR Master Mix | G3327-01 | 1 mL |
| G3327-05 | 5×1 mL | |
| G3327-15 | 15×1 mL |
Product Description
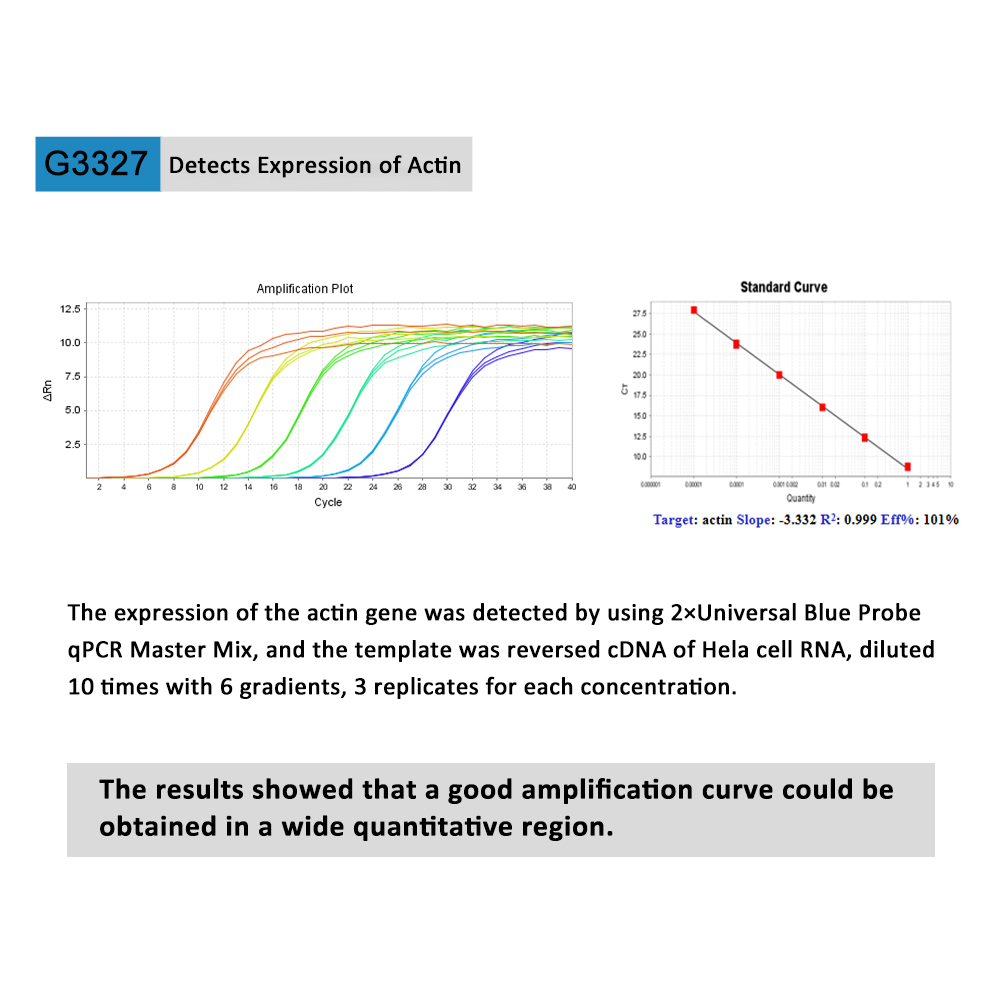
The 2×Universal Blue Probe qPCR Master Mix is a 2X reaction mix optimized for real-time qPCR detection and quantitation of target DNA sequences using hydrolysis probes. It contains Hot Start Taq DNA polymerase and a reaction buffer optimized for Probe qPCR to ensures a good amplification curve, accurate quantification of target genes ,increased PCR specificity and sensitivity, good repeatability and high reliability over a broad dynamic range. Only templates, primers, probes and Nuclease-free Water need to be added for use. It also features a unique passive reference dye that is compatible across a variety of instrument platforms and a non-fluorescent, visible dye to monitor reaction setup. This dye does not spectrally overlap fluorophores commonly used for qPCR and will not interfere with real-time detection.
Storage and Shipping Conditions
Ship with wet ice; store at -20°C away form light, valid for 12 months..
Product Contents
| Component Number | Component | G3327-01 | G3327-05 | G3327-15 |
| G3327-1 | 2×Universal Blue Probe qPCR Master Mix | 1 mL | 5×1 mL | 15×1 mL |
Assay Protocol
Before starting
1. Real-Time PCR Instruments;
2. PCR tubes, PCR plates or accessories offer excellent qPCR performance;
3. PCR primers (reference primer design principles);
Procedures
1. Gently vortex and briefly centrifuge all solutions after thawing.
2. Prepare a reaction master mix by adding the following components (except template DNA) for each reaction to a tube on ice:
| Component | 20 μL rxn | 50 μL rxn | Final Concentration |
| 2×Universal Blue Probe qPCR Master Mix | 10 μL | 25 μL | 1× |
| Forward Primer (10 μM)a | 0.4μL | 1 μL | 0.2 μM |
| Reverse Primer (10 μM)a | 0.4μL | 1 μL | 0.2 μM |
| Probe (10 μM) a | 0.4μL | 1 μL | 0.2 μM |
| Template b | Variable | Variable | as required |
| Nuclease-free Water | Add to 20 μL | Add to 50 μL |
a. Usually, a good amplification effect can be obtained with the final concentration of 0.2 μM. When the reaction performance is poor, the primer concentration can be adjusted in the range of 0.2-1.0 μM.
b. The amount of template added varies depending on the number of copies of the target gene, and the appropriate amount of template addition is studied by gradient dilution. The best addition amount of template DNA in the 20 μL reaction system was less than 100 ng. When the cDNA (RT reaction solution) of RT-PCR reaction was used as template, the addition amount should not exceed 10% of the final qPCR volume.
3. Mix the master mix thoroughly and dispense appropriate volumes into PCR tubes or plates.
4. Add template DNA to the individual PCR tubes or wells containing the master mix.
5. Gently mix the reactions without creating bubbles (do not vortex). Centrifuge briefly if needed. Bubbles will interfere with fluorescence detection.
6. Program the thermal cycler according to the recommendations below, place the samples in the cycler and start the program.
| A. Two-step method | B. The three-step method | ||||||||
| stage | step | Cycle number | temperature | time | stage | step | Cycle number | temperature | time |
| Stage 1 | Predegeneration | 1 | 95℃ | 30 sec | Stage 1 | Predegeneration | 1 | 95℃ | 30 sec |
| Stage 2 | Denaturation | 40 | 95℃ | 15 sec | Stage 2 | Denaturation | 40 | 95℃ | 15 sec |
| Annealing
/extension |
60℃ | 30 sec | Annealing | 55-65℃ | 10 sec | ||||
| Extension | 72℃ | 30 sec | |||||||
a: If amplification specificity needs to be improved, two-step procedure or annealing temperature can be used; To improve the amplification efficiency, a three-step procedure or extension time can be used.
Note
1. Mixed gently upside down before use. Do not swirl and shake to creat bubbles.
2. Reagents should be placed on ice when preparing reaction mixes.
3. Strong light should be avoided when preparing PCR reaction mixes due to fluorescent dye SYBR Green.
4. New disposable tips should be used for preparation of reaction mixes to avoid cross contamination.
5. Avoid freeze-thawing cycles of the Master Mix and try to use it up within one month after thawing.
Compatible instruments
ABI: 5700, 7000, 7300, 7700, 7900, 7900HT, 7900 HT Fast, StepOne™, StepOne Plus™, 7500/7500 Fast, ViiA 7™, QuantStudio™ 系列, PikoRealTM Cycler;
Stratagene: Mx3000P®, 3005P™, 4000™;
Bio-Rad: CFX96™, CFX384™, iCycler iQ™, iQ5™, MyiQ™, MiniOpticon™, Opticon®, Opticon 2, Chromo4™;
Eppendorf: Realplex 2s, Mastercycler® ep, realplex;
IIIumina: Eco QPCR;
Cepheid: SmartCycler®;
Qiagen Corbett: Rotor-Gene® series.
Roche: LightCycler™ Series;
Takara: Thermal Cycler Dice Series;
Analytikjena: qTOWER Series;
Hangzhou Bori: LineGene Series.
Taqman primer design Principles
1. Determine the probe first, then design the primer;
2. When designing primers, get as close to the probe as possible without overlapping the probe;
3. Avoid runs of identical nucleotides. If repeats are present, there must be fewer than four consecutive G residues;
4. Optimal melting temperature (Tm): 60°C. Differences in Tm of the two primers should not exceed 2°C;
5. Avoid more than two G or C nucleotides in the last five nucleotides at 3′ end to lower the risk of nonspecific priming.
6. Avoid secondary structures in the amplicon.
7.Avoid self-complementarities in a primer, complementarities between the primers and direct repeats in a primer to prevent hairpin formation and primer dimerization.
8. In order to avoid genome amplification, primers should preferably be designed across exons;
9. The Optimal amplicon length should be 50-150bp to obtain the best PCR efficiency;
9. No other non-specific products appeared on NCBI blast.
Taqman probe design Principles
1. The probe length should be 13-25bp(13-30bp if conventional TaqMan probe is used);
2. The Tm value should be 65°C~70°C, usually 5~10℃ higher than the Tm value of the primer, to ensure that the probe is preferentially bound to the target gene during the annealing process;
3. Aim for the GC content to be between 40 and 70%, with the 3′ of a primer ending in C or G to promote binding.
4. Exclude G at the 5′ end of the probe, which causes quenching.
5. Select the template DNA strand containing more C than G bases.
6. Avoid secondary structures.
7.Avoid dimerization with primers
Taqman MGB Probe Design Principles
1.A reporter dye (e.g., FAM™ dye) is attached to the 5 ‘end of the probe;
2. There is a non-fluorescent quencher (NFQ) at the 3 ‘end of the probe;
3. The MGB moiety stabilizes the hybridized probe and effectively raises the melting temperature (Tm). This means MGB™ probes can be shorter than traditional dual-labeled probes, not less than 13 bp;
4. The MGB probe can provide better sequence discrimination even point mutation(the MGB probe is not bind to the target gene and fluorescence signal do not appear).
For Research Use Only!
|
Cat.No.
|
Product Name
|
Spec.
|
Operation
|
|---|
|
G3326-01
|
2×Universal Blue SYBR Green qPCR Master Mix With 40×Yellow Template Dilution Buffer
|
1 mL
|
|
|
G3326-05
|
2×Universal Blue SYBR Green qPCR Master Mix With 40×Yellow Template Dilution Buffer
|
5×1 mL
|
|
|
G3326-15
|
2×Universal Blue SYBR Green qPCR Master Mix With 40×Yellow Template Dilution Buffer
|
15×1 mL
|
|
|
G3327-01
|
2×Universal Blue Probe qPCR Master Mix
|
1 mL
|
|
|
G3327-05
|
2×Universal Blue Probe qPCR Master Mix
|
5×1 mL
|
|
|
G3327-15
|
2×Universal Blue Probe qPCR Master Mix
|
15×1 mL
|
|
|
G3328-01
|
2×Universal Blue SYBR Green qPCR Master Mix (with UDG)
|
1 mL
|
|
|
G3328-05
|
2×Universal Blue SYBR Green qPCR Master Mix (with UDG)
|
5×1 mL
|
|
|
G3328-15
|
2×Universal Blue SYBR Green qPCR Master Mix (with UDG)
|
15×1 mL
|